
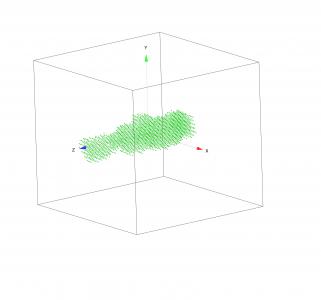
Read PDB files and display in Gizmo
This code will load PDB files as described here:
https://en.wikipedia.org/wiki/Protein_Data_Bank_(file_format)
and display them in Gizmo. There are many software packages able to display PDB files, but most are large and bit challenge to use. This code snippet will import PDB file, extract atom positions and display them in Gizmo.
//This set of procedures will read PDB (protein database) files: // https://en.wikipedia.org/wiki/Protein_Data_Bank_(file_format) // //Jan Ilavsky, 2018 //free to use or modify, no copyright. // Major thanks to John Weeks who wrote most of early code when I needed it // Thanks to tcaus, Igor Exchange user who described the pdb file structure: // referecne: https://www.wavemetrics.com/forum/general/igor-does-not-recognize-decimal-numbers-my-data //To use: // run ReadPDBFile() in command line. Function ReadPDBFile() // the following pdb description is from Igor user group user icaus, original reference available here: // https://www.wavemetrics.com/forum/general/igor-does-not-recognize-decimal-numbers-my-data // // start in known place... String NewFoldername setDataFolder root: //this is definition of way PDB locations are written //note that most of data I do not need, you may need to change to code to read and use atoom names and other info. String columnInfo = "" // This only works for rows starting with "ATOM" columnInfo+="C=1,W=6,N=keyword;" //"ATOM" name column 0 columnInfo+="C=1,W=6,N='_skip_';" // Atom number (last column unspecified) 7 columnInfo+="C=1,W=5,N='_skip_';" // Atom name; last character is "alt Loc" 13 columnInfo+="C=1,W=3,N='_skip_';" // Residue name; starts at column 18 18 columnInfo+="C=1,W=2,N='_skip_';" // Chain identifier in column 22 (21=?) 21 columnInfo+="C=1,W=4,N='_skip_';" // Residue sequence number 23 columnInfo+="C=1,W=4,N='_skip_';" // Blank stuff 27 columnInfo+="C=1,W=8,N=xval;" // Should start at column 31 31 columnInfo+="C=1,W=8,N=yval;" // 39 columnInfo+="C=1,W=8,N=zval;" // 47 columnInfo+="C=1,W=6,N='_skip_';" // Occupancy columnInfo+="C=1,W=6,N='_skip_';" // Temperature info columnInfo+="C=1,W=11,N='_skip_';" // Element symbol (col 77,78) columnInfo+="C=1,W=2,N='_skip_';" // Charge (col 79,80) //OK, now load the data... LoadWave/A/O/F={14,8,0}/K=2/B=columnInfo Wave/T xval, yval, zval Wave/T keyword //create name for future data location, unique name created from loaded file name. NewFoldername= UniqueName(StringFromList(0,S_FileName,"."), 11, 0) //remove all non ATOM lines from the waves... Variable count=numpnts(keyword)-1 do if (strsearch(keyword[count], "ATOM", 0) <0 ) DeletePoints count, 1, keyword, xval, yval, zval endif count-=1 while (count >= 0 ) //create location for centers of atoms in Gizmo and assign the positions. Make/O/D/n=(numpnts(xval),3) centersWave=nan centersWave[][0]=str2num(xval[p]) centersWave[][1]=str2num(yval[p]) centersWave[][2]=str2num(zval[p]) //cleanup KillWaves keyword, xval, yval, zval //create location for data NewDataFolder/S $(NewFoldername) //move there the centers wave MoveWave root:centersWave, $(GetDataFolder(1)) //now create the Gizmo, it needs to know where the data are, this is one way to tell it... string PathToData=GetDataFolder(1)+"centersWave" MakeGizmo(PathToData) End Function MakeGizmo(PathToData) string PathToData //will create Gizmo, I used existiing Gizmo and created recreation function. //these are the centers... Wave GizmoData=$(PathToData) NewGizmo/K=1/W=(83,132,981,947) ModifyGizmo startRecMacro=700 ModifyGizmo scalingMode=8 ModifyGizmo scalingOption=0 AppendToGizmo Scatter=GizmoData,name=scatter0 ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ scatterColorType,0} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ markerType,0} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ sizeType,0} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ rotationType,0} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ Shape,1} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ size,1} ModifyGizmo ModifyObject=scatter0,objectType=scatter,property={ color,4.57771e-05,0.8,1.5259e-05,1} AppendToGizmo Axes=boxAxes,name=axes0 ModifyGizmo ModifyObject=axes0,objectType=Axes,property={-1,axisScalingMode,1} ModifyGizmo ModifyObject=axes0,objectType=Axes,property={-1,axisColor,0,0,0,1} ModifyGizmo modifyObject=axes0,objectType=Axes,property={-1,Clipped,0} AppendToGizmo attribute pointSize=8, name=pointSize0 ModifyGizmo setDisplayList=0, attribute=pointSize0 ModifyGizmo setDisplayList=1, object=scatter0 ModifyGizmo setDisplayList=2, object=axes0 ModifyGizmo currentGroupObject="" ShowTools ModifyGizmo showInfo ModifyGizmo infoWindow={904,365,1721,662} ModifyGizmo showAxisCue=1 ModifyGizmo pan={0.075688,-0.110429} ModifyGizmo idleEventQuaternion={-1.1184e-06,6.7395e-06,-4.52601e-06,1} //these next two lines scale the Gizmo axes to be 5% larger than Max atom position value. //*1.05 should make sure the all positions are inside, not clipped, and x,y, and z have same scale //(so dimensions are not distorted). WaveStats/Q GizmoData ModifyGizmo setOuterBox={-1.05*V_max,1.05*V_max,-1.05*V_max,1.05*V_max,-1.05*V_max,1.05*V_max} end

Forum

Support
Gallery
Igor Pro 10
Learn More
Igor XOP Toolkit
Learn More
Igor NIDAQ Tools MX
Learn More
