
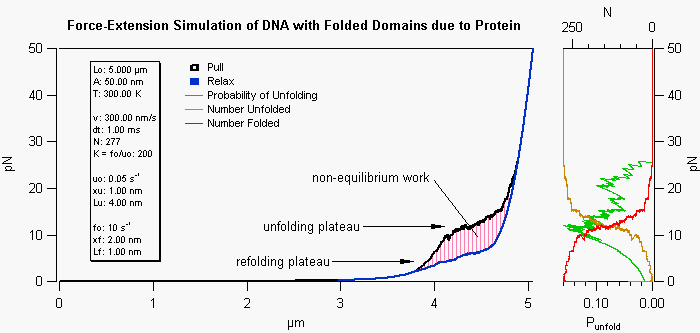
This simulation procedure uses an extensible worm-like-chain (WLC) model to depict the force-extension behavior of DNA with or without distorting proteins. Here the proteins are assumed to fold the DNA in some manner to reduce its normal contour length. Parameters defining the energy well of each folded domain as well as the properties of the naked and protein covered DNA are entered in the panel. The amount of bound protein is decided by the protein dissociation constant and concentration. As the DNA is extended the force increases according to the WLC model and the barrier to unfolding is decreased. When an unfolding event is reached the contour length of the DNA is extended and the unfolded segment of DNA is described by a completely separate group of parameters. Thus the extension of the DNA/protein complex retains memory of how many domains have been unfolded by including additional characteristics for the unfolded, protein bound, segments of the DNA. The hysteresis in the pullelax cycle occurs if the pulling rate is faster than the equilibrium rate of folding/unfolding events. The plots on the right show the probability of an unfolding event, as well as, the number of unfolded/folded versus force.
Raymond W. Friddle, Lawrence Livermore National Laboratory

Forum

Support
Gallery
Igor Pro 10
Learn More
Igor XOP Toolkit
Learn More
Igor NIDAQ Tools MX
Learn More





