
VisualEnzymics Interface
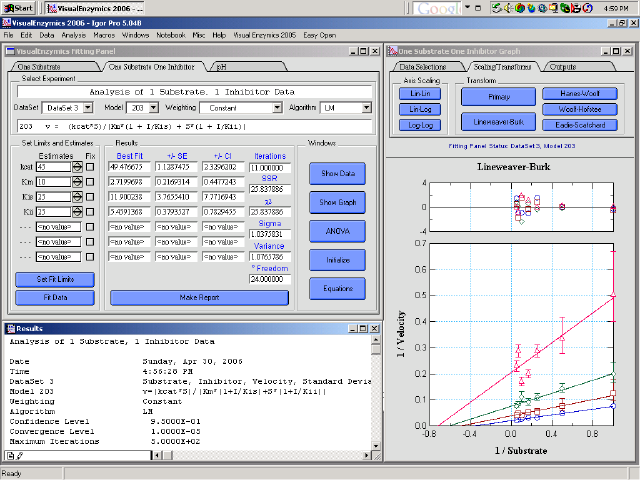
Click Here for a large screen shot from a Macintosh.
VisualEnzymics is a software package for the analysis of enzyme kinetic data. The program was developed in Igor Pro and consists of several modules for fitting substrate saturation data, enzyme inhibition data, and enzyme pH variation data.
The VisualEnzymics interface promotes interactive data analysis where the user can fit different kinetic models to a single dataset and see the results instantly. The plots can be transformed interactively to view Lineweaver-Burk, Hanes-Woolf, Woolf-Hofstee, and Eadie-Scatchard plots. Fits to different kinetic models can be compared in an ANOVA module.
Preformatted layouts can be generated with a single button click. Statistical reports can be generated in a notebook window. The program uses the FuncFit operation for model fitting, and VisualEnzymics has added a Monte Carlo routine for brute force fitting when it is hard to find good initial estimates. VisualEnzymics also has added proportional weighting for data that varies over several orders of magnitude. The program currently offers 35 kinetic models for analysis.
Additional features can be viewed at www.softzymics.com.
Submitted by
James G. Robertson
SoftZymics, Inc.
Makers of VisualEnzymics for analysis of enzyme kinetic data

Forum

Support
Gallery
Igor Pro 9
Learn More
Igor XOP Toolkit
Learn More
Igor NIDAQ Tools MX
Learn More





